PUBLICATIONS > Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine
7 June 2013
Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine
Authors: Zuanel Diaz, Adriana Aguilar-Mahecha, Eric R Paquet, Mark Basik, Michèle Orain, Errol Camlioglu, André Constantin, Naciba Benlimame, Dimcho Bachvarov, Guillaume Jannot, Martin J Simard, Benoit Chabot, Adrian Gologan, Roscoe Klinck, Thérèse Gagnon-Kugler, Bernard Lespérance, Benoit Samson, Petr Kavan, Thierry Alcindor, Richard Dalfen, Cathy Lan, Catherine Chabot, Marguerite Buchanan, Ewa Przybytkowski, Samia Qureshi, Caroline Rousseau, Alan Spatz, Bernard Têtu & Gerald Batist
Abstract
Great advances in analytical technology coupled with accelerated new drug development and growing understanding of biological challenges, such as tumor heterogeneity, have required a change in the focus for biobanking. Most current banks contain samples of primary tumors, but linking molecular signatures to therapeutic questions requires serial biopsies in the setting of metastatic disease, next-generation of biobanking. Furthermore, an integration of multidimensional analysis of various molecular components, that is, RNA, DNA, methylome, microRNAome and post-translational modifications of the proteome, is necessary for a comprehensive view of a tumor’s biology. While data using such biopsies are now regularly presented, the preanalytical variables in tissue procurement and processing in multicenter studies are seldom detailed and therefore are difficult to duplicate or standardize across sites and across studies. In the context of a biopsy-driven clinical trial, we generated a detailed protocol that includes morphological evaluation and isolation of high-quality nucleic acids from small needle core biopsies obtained from liver metastases. The protocol supports stable shipping of samples to a central laboratory, where biopsies are subsequently embedded in support media. Designated pathologists must evaluate all biopsies for tumor content and macrodissection can be performed if necessary to meet our criteria of >60% neoplastic cells and <20% necrosis for genomic isolation. We validated our protocol in 40 patients who participated in a biopsy-driven study of therapeutic resistance in metastatic colorectal cancer. To ensure that our protocol was compatible with multiplex discovery platforms and that no component of the processing interfered with downstream enzymatic reactions, we performed array comparative genomic hybridization, methylation profiling, microRNA profiling, splicing variant analysis and gene expression profiling using genomic material isolated from liver biopsy cores. Our standard operating procedures for next-generation biobanking can be applied widely in multiple settings, including multicentered and international biopsy-driven trials.
Main
As significant strides are being made in individualized therapy, increasing emphasis is being placed on obtaining serial biopsies from the same individual before therapy and at later time points, to explore discordances between primary tumors and metastases, gain insight into the mode of action of an investigational agent, elucidate resistance mechanisms and ultimately to guide therapy.1, 2, 3 Adaptive clinical trials such as the Biomarker-Integrated Approaches of Targeted Therapy for Lung Cancer Elimination (BATTLE) trial and the Investigation of Serial Studies to Predict Your Therapeutic Response with Imaging and Molecular Analysis 2 (I-SPY2) trial in breast cancer are recent examples of the use of biopsies to evaluate the molecular composition of evolving tumors and to detect biomarkers for patient stratification.4, 5
There is no universal method for biospecimen processing since this differs depending on the demands of the analytical platforms. Some journals now require key preanalytical variables to be reported, according to the framework provided by the Biospecimen Reporting for Improved Study Quality (BRISQ) guidelines.6, 7 For multiplexed downstream analyses of small and fragile specimens such as needle core biopsies, standardized procedures are especially important to obtain high-quality genomic material with reasonable yield. However, histomorphological quality control is essential before pursuing unbiased discovery approaches since assumptions on tumor cellularity can skew results, as can preanalytical variables in tissue processing, including procurement (biospecimen excision method, warm and cold ischemic time), preservation (snap freezing or use of stabilization reagents, preservation temperature, freeze and thaw cycles, shipping temperature), embedding (type of long-term preservative, storage parameters) and nucleic acid extraction methods.8, 9, 10 These variables may contribute to the lack of reproducibility of major publications in biomarker discovery.11
Our objective was to develop a standardized method that would allow for the analysis of tumor content and extraction of high-quality DNA and RNA with sufficient yields for unbiased discovery platforms, from a single liver needle core biopsy. We evaluated different tissue procurement methods and embedding media, and developed standard operating procedures for use in a multicenter study involving the collection of liver biopsies from metastatic colorectal cancer patients (study Q-CROC-01). We demonstrate here that our next-generation biobanking protocol is compatible with downstream applications, including array comparative genomic hybridization, methylation profiling, microRNA profiling, splicing variant analysis and gene expression profiling.
Materials and methods
Tissue Sampling from Surgical Specimens
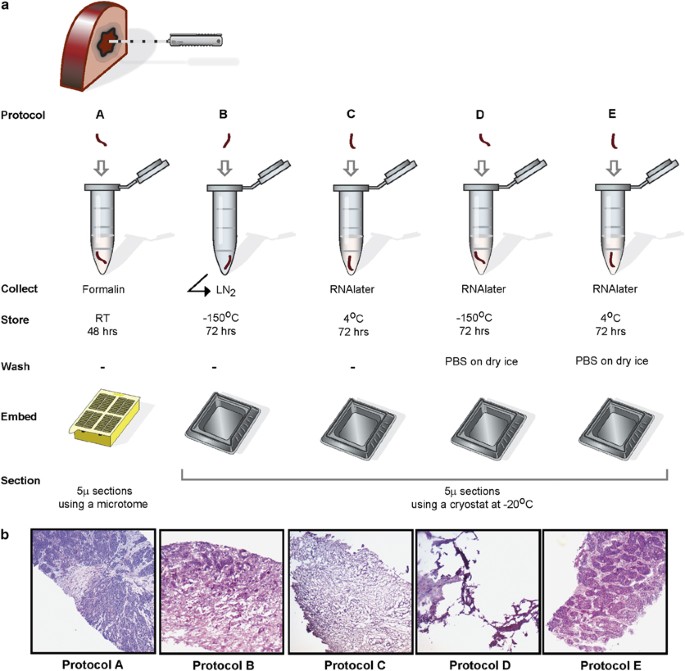
Patients provided written consent according to the Research Ethics Board of the Jewish General Hospital. Biopsies were taken from two different hepatectomy samples using a 16 G BioPince full-core end-cutting tri-axial automated biopsy device with a stroke length set to 23 mm. The biopsy cores were approximately 1 mm × 15 mm. A minimum of six biopsy cores were processed for each protocol detailed in Figure 1. Standard pathology procedures were followed for the formalin-fixed, paraffin-embedded sample (protocol A). For protocol B, the biopsies were collected in an empty microcentrifuge tube, immediately submerged in liquid nitrogen and then stored at −150 °C for 72 h. For protocol C, biopsies were immediately submerged in 150 μl RNAlater solution (Qiagen, Ontario,Canada) and then stored at 4 °C for 72 h. For protocols D and E, biopsies were submerged in 150 μl RNAlater and then stored at −150 °C or 4 °C respectively, for 72 h. Frozen samples from protocol D were thawed gently, kept on ice and, along with samples from protocol E, were washed with RNAse-free phosphate-buffered saline (PBS) (Fisher Scientific, Ottawa, ON, Canada) for 2 min on dry ice and repeated two times. Biopsies from protocols B, C, D and E were embedded in optimal cutting temperature (OCT) compound (Surgipath; Leicabiosystems, Concord, ON, Canada) using a 15 mm × 15 mm Tissue Tek disposable cryomold (Somagen, Edmonton, AB, Canada). Cryomolds were gently submerged for 30 s into a beaker containing 2-methyl butane (Fisher Scientific) precooled on dry ice. The blocks were stored at −80 °C. Tissue cryosections were performed using standard procedures, with the cryostat temperature set to −20 °C.
Tissue Sampling from Colorectal Cancer Patients
Written informed consent from patients was obtained before all study-related procedures, which were approved by the ethics committee of five hospitals. Prebiopsy coagulation status was assessed according to the Society of Interventional Radiology Clinical Practice Guideline.12 Interventional radiologists specifically aimed at including 100% tumor tissue in the biopsy. Three cores were removed by percutaneous ultrasound-guided biopsies using primarily a 16- or 18-G BioPince end-cutting biopsy device. After the biopsy procedure, patients were monitored for 4–6 h by nursing staff. Standard operating procedures, including needle core biopsy procurement, histological analysis, isolation, quantitation and quality control of genomic material, are available as Supplementary Material.
Array Comparative Genomic Hybridization
Copy number alterations within the tumor genome relative to the commercially available normal human DNA (Promega, Madison, WI, USA) were identified by array comparative genomic hybridization analysis using 244K Agilent platform (Agilent Technologies, Mississauga, ON, Canada). We used whole genome amplification procedure before labeling and hybridization to the array, as reported previously.13 Thus, only 60 ng of patient samples and reference DNA was required. Whole genomic DNA was amplified using GenomiPhi V2 DNA Amplification Kit (GE Healthcare, Baie d’Urfe, QC, Canada). Amplified DNA was labeled and purified according to the protocol supplied by Agilent. Data were extracted from scanned images using the Feature Extraction software, version 10.10.1.1 (Agilent Technologies) and analyzed using aberration detection method 2 (ADM-2) algorithm to identify DNA copy number aberrations.13 The text files from Feature Extraction were also imported for analysis into R using the package limma and the processed signal provided by Feature Extraction was used for downstream visualization.14 The data were visualized using the Broad Institute Integrative Genomics Viewer.15
Methylation Profiling
Global methylation profiling was performed using the methylated DNA immunoprecipitation approach coupled with detection of methylated regions on high-density (244K) Agilent human CpG island arrays that interrogate 195K CpG island probes and 50K non-CpG island probes (Agilent Technologies). For methylated DNA immunoprecipitation, we used the Agilent protocol version 1.1 without any modifications. Microarray slides were scanned using the Agilent G2505B scanner and data extracted with Agilent Feature Extraction 9.1 software. We used the package limma to read and preprocess the raw data. Briefly, we subtracted the background and normalized using the median. Significant hypomethylated or hypermethylated CpG regions were identified by comparing the log 2 ratio of probes within a CpG in a Wilcoxon’s signed-rank test. Regions with a P-value lower than 0.0001 were considered as significant. The data were visualized using the Broad Institute Integrative Genomics Viewer.15
Splicing Isoforms
Analysis of alternative splicing events from total RNA was performed using a high-throughput RT-PCR approach.16, 17 RNA (25 ng) extracted from the biopsies was amplified using the Transplex Whole Transcriptome Amplification Kit (Sigma-Aldrich, Oakville, ON, Canada). Relative expression levels of a panel of reference genes were determined in each sample by qPCR to normalize the amplification of all RNA samples analyzed. Nine reference genes were amplified (YWHAZ, GAPDH, SDHA, HMBS, SF3A1, HPRT1, PUM1, MRPL19 and PSMC4, primer sequences available on request), and the three most stable between samples (SDHA, HMBS and PSMC4) were identified using the GeNorm protocol.18 Then, the samples were diluted accordingly to the lowest concentration of the sample set, then further diluted 10-fold, for subsequent high-throughput PCR analysis of alternative splicing. End point PCR followed by microcapillary electrophoresis, targeting 96 alternative splicing events known to be active in a panel of 11 cell lines (unpublished data), were analyzed in the three amplified samples as described previously.16 The full data set for the normalization and alternative splicing analysis is available at http://palace.lgfus.ca/link/qcroc.
MicroRNA Profiling
MicroRNA profiling was performed using TaqMan low-density arrays according to the manufacturer’s protocol (Life Technologies, Carlsbad, CA, USA) using 100 ng of total RNAs, and quantified using the RQ manager 1.2.1 software (Life Technologies). No additional step was applied to the RNA extraction to enrich for microRNAs. Text files were imported using a custom script within R (http://www.r-project.org/). Data were normalized using the mean snRNA U6 Ct.
Gene Expression Profiling
Microarray analysis was performed using 100 ng of RNA, which was amplified and labeled according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies). Samples were hybridized to SurePrint G3 Human GE 8 × 60K array slides (Agilent Technologies). Slides were processed and scanned according to the manufacturer’s instructions. Data were extracted using Feature Extraction software, version 10.10.1.1 (Agilent Technologies) and imported in R using BioConductor (http://www.bioconductor.org/) and the package limma.14 We used Pearson’s correlation to assess the level of correlation between the gene expression experiments. For data integration between gene expression and array comparative genomic hybridization, we first scaled the data. To detect the concordance of the level of expression with deletions (or amplifications) identified by array comparative genomic hybridization, we defined background regions without chromosomal aberrations from ADM-2 results and then compared the distribution of the scaled expression within the background region to the region of interest. The significant difference in scaled expression distribution for the background region and the deleted (or amplified) region was evaluated using a Wilcoxon’s signed-rank test.
Results
Optimizing Conditions for Histological Analysis
To determine conditions that permit morphology analysis and ensure nucleic acid integrity, we designed a pilot study using cores from a fresh surgically resected liver of a patient with metastatic colorectal cancer. Figure 1 demonstrates the preanalytical workflow from collection to histological verification, testing three conditions for specimen collection: formalin, snap freezing in liquid nitrogen and RNAlater. To minimize the dehydration effect of RNAlater, we washed some biopsies with ice-cold PBS (protocols D and E). All biopsies were embedded in OCT compound to enable cryosectioning, with the exception of the biopsies collected in formalin (protocol A), which were routinely processed and paraffin embedded. As expected, optimal morphological preservation was observed in these samples (Figure 1b). Biopsies snap frozen in liquid nitrogen (protocol B) also exhibited adequate morphology but specimens were fragile and more difficult to section. Insufficient histology preservation was observed for samples processed according to protocols C and D.19, 20 When the samples were frozen in RNAlater (protocol D), the tissue did not adhere to the slide properly and staining was only observed in the periphery or in distant fragments of the tissue, rendering histological examination impossible, despite the additional washing step. As protocols C and D did not result in suitable morphology, which was a technical requirement of our method, we did not pursue any recovery of DNA and RNA from these samples. Suitable morphology was only observed when samples in RNAlater were stored at 4 °C and washed with ice-cold PBS before OCT embedding (protocol E).
Recovery of Genomic Material
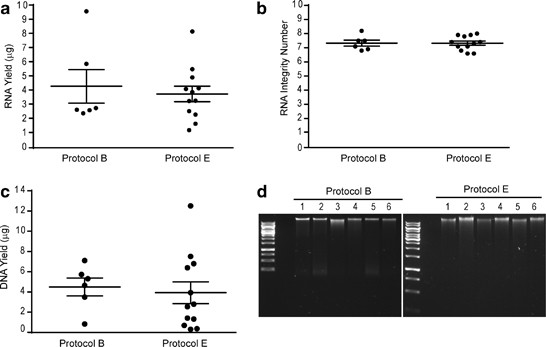
The main concern in isolating intact genomic material suitable for downstream expression analyses is the fragile nature of RNA and its susceptibility to degradation. To enable array comparative genomic hybridization, methylation, gene expression, microRNA and splicing variant profiling, we required a minimum yield of 1.06 μg DNA and 0.225 μg RNA. We used the Qiagen AllPrep DNA/RNA Mini Kit to isolate genomic material from the biopsies processed by protocols B and E, thereby allowing us to isolate simultaneously DNA and RNA from the same biopsy. Formalin-fixed, paraffin-embedded samples (protocol A) were not selected for further processing since historically these are not optimal for ‘-omics’ technologies due to crosslinking and fragmentation.21 As shown in Figure 2a, both protocols B and E provided good RNA yields, with a mean of 4.28 μg for biopsies snap frozen in liquid nitrogen and 3.71 μg for biopsies collected in RNAlater. The ratio of the absorbance at 260 and 280 nm (A260/280) was used to assess the purity of the nucleic acids, and both protocols yielded values >1.8 for DNA and 2 for RNA, indicating minimal contamination (data not shown). All samples processed according to protocols B and E had RNA integrity numbers above 6.8, with a mean of 7.3 (Figure 2b). The mean DNA isolated from the same biopsies was 4.5 μg for snap frozen samples and 3.9 μg for samples collected in RNAlater (Figure 2c). We observe more variability in DNA yield with some samples having very low yields, which we attribute to suboptimal homogenization of the tissue. We have noticed that RNAlater makes the tissue harder and more difficult to homogenize. High-molecular-weight bands were detected for all samples by agarose gel electrophoresis, thereby confirming DNA integrity (Figure 2d).
Validation in a Multicenter Study
The Q-CROC-01 study aims to identify prospectively biomarkers of clinical resistance to first-line therapy in metastatic colorectal cancer. To facilitate the procurement, transport and manipulation of liver biopsies from multiple centers participating in this study, collection in RNAlater was selected. Three needle core biopsies were collected before first-line treatment; the first two biopsies (Biopsy-1 and Biopsy-2) were placed in RNAlater for isolation of genomic material and the third (Biopsy-3) was collected in formalin. Biopsies were shipped overnight at 4 °C to our central laboratory for all subsequent sample processing. The two biopsies in RNAlater were processed using protocol E. A designated pathologist evaluated all first and second biopsies, noting tumor content in the entire core. We defined tumor content as neoplastic cells (tumor nuclei), necrosis and stroma. The pathologist first marked on the H&E slide if macrodissection of the biopsy was necessary and feasible, and then further characterized the marked tumor-containing region, noting the percentage of neoplastic cells, necrotic cells and stroma. Although there is no consensus on the minimum tumor content required for reliable analysis from high-throughput platforms, we adopted the requirement defined by the Cancer Genome Atlas (TCGA) for genome-scale characterization of human colon and rectal cancer.22 Thus, within the marked tumor section, only samples containing >60% neoplastic cells and <20% necrosis met our criteria for genomic isolation. All samples not meeting these criteria were excluded, and genomic material was not isolated.
Of the first 42 patients enrolled into the Q-CROC-01 study at five centers, two did not undergo the liver biopsy procedure. As shown in Figure 3a, of the 40 patients who were biopsied before starting first-line treatment, three patients had biopsy samples that were not histologically consistent with colorectal cancer. Of the 37 patients whose biopsies were consistent with colorectal cancer, 29 (78%) had at least one biopsy meeting our criteria for downstream DNA and RNA isolation. Eight patients failed quality control in both biopsies, and subsequently no DNA or RNA was isolated; seven patients had insufficient neoplastic cells (<60%) in both biopsies 1 and 2 with no potential for macrodissection, and for one patient, the site erroneously placed all biopsies in formalin.
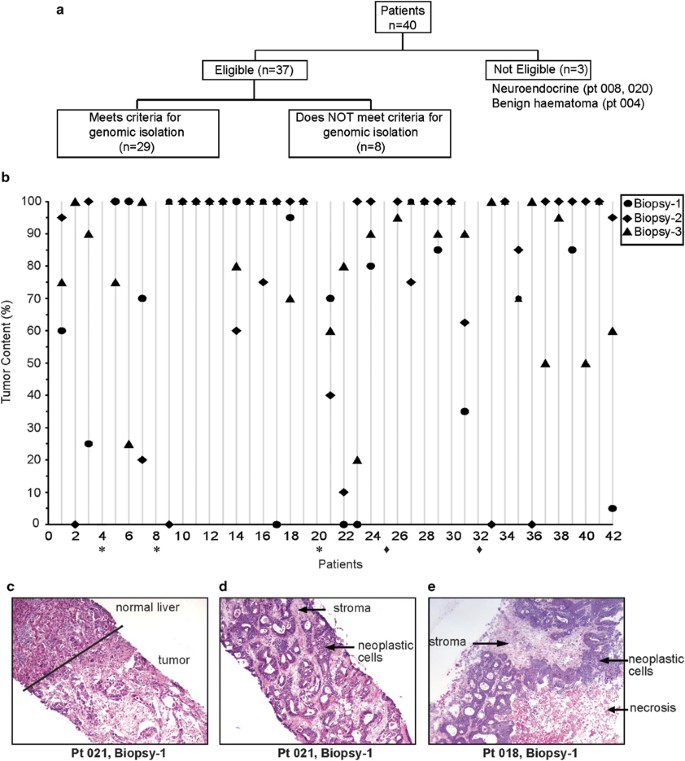
As shown in Figure 3b, there is significant variability in the tumor content between biopsies and even between three successive biopsy passes taken from the same patient. Even biopsies containing 100% tumor (includes neoplastic cells, necrotic cells and stroma) may not have sufficient neoplastic cells to warrant genomic material isolation as defined by our criteria. For example, biopsies 1 and 2 from patient 30 consisted entirely of tumor tissue, but as the cores contained primarily necrotic cells and stroma, the samples were not suitable for isolation of genomic material. Figure 3c demonstrates the boundary between normal and tumor tissue in liver biopsy 1 from patient 021, in which macrodissection was recommended. Macrodissection was performed on dry ice with an RNAse-free sterile blade, using the H&E slide as a reference to cut the OCT block in the region delineated by the pathologist (area containing neoplastic cells with little stroma or necrosis). Only the macrodissected tissue containing the area of interest was then used to isolate genomic material (Figure 3d). In some biopsies, there was no clear delimitation that could be made to enrich for neoplastic cells (Figure 3e). No correlation could be made at this point between the variability in tumor content between biopsies and the five centers at which the biopsies were performed.
From the biopsies that met our criteria for genomic isolation, variable yields of DNA and RNA were obtained, ranging between 1.0 and 27.8 μg for DNA (mean of 12.6 μg) and between 0.89 and 17.21 μg for RNA (mean of 7.9 μg). High purity was obtained with A260/280 values >1.8 for DNA and 2 for RNA, confirming the validity of the protocol in patient liver samples. The quality of the RNA and DNA was also verified and RNA integrity numbers higher than 6 (average=7.1) were obtained for RNA and intact genomic DNA was observed (data not shown).
Validation for Downstream High-Throughput Technologies
To ensure that our next-generation biobanking protocol is compatible with multiplex discovery platforms and that no component of the sample preparation interferes with downstream enzymatic reactions, we performed array comparative genomic hybridization, DNA methylation, microRNA, splice variant and gene expression profiling, using three patient samples with the highest yield of DNA and RNA. The rest of the samples were preserved for ‘batch analyses’ at the end of the study.
Array comparative genomic hybridization
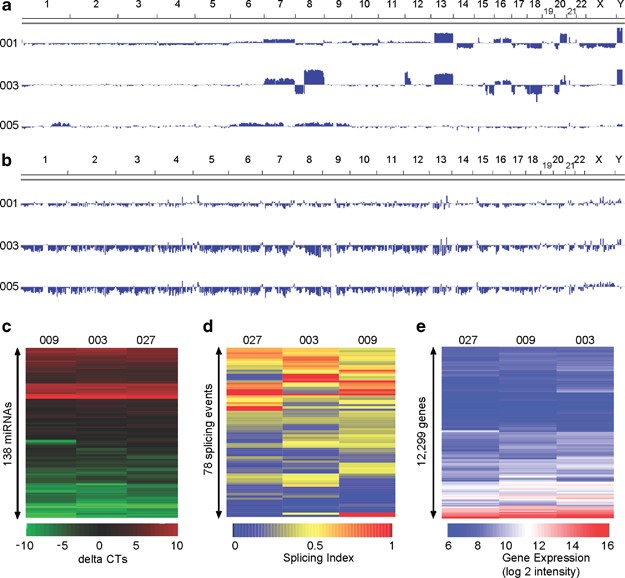
Patient biopsy samples were analyzed for DNA copy number changes using Agilent’s Genomic Workbench software ADM-2. No aberrations were identified for patient 005, but patients 001 and 003 showed multiple aberrations that mostly involved large DNA segments (Figure 4a). Many of these genetic aberrations occurred in regions reported to be associated with colorectal cancer.23, 24 These changes include amplifications on chromosome 20q, 13 and 8q, and copy number loss on the long arm of chromosome 18 and on chromosome 8p.24, 25, 26, 27, 28 Both aberrant specimens showed amplifications of entire chromosomes: 7, 13 and 16, as well as partially overlapping aberrations on chromosome 20 (Figure 4a).
Methylation profiling
To enable genome-wide analysis of methylation status of known CpG islands, we used an affinity-based approach to enrich for methylated DNA, and evaluated methylation status using Agilent Human DNA Methylation microarray. As shown in Figure 4b, we observed global hypomethylation pattern, mainly negative values, for patients 003 and 005. Global hypomethylation of DNA is observed frequently during carcinogenesis and has been reported in colorectal cancer.29, 30 We analyzed the methylation profile on the genes implicated in colorectal cancer as reported by the Kyoto Encyclopedia of Genes and Genomes.31 We observed hypomethylation of CpG islands of JUN (P<3.1e−06), FOS (P<9.5e−4) and MYC (P<3.8e−04), as shown in Figure 5c. MYC is reported to be hypomethylated in several colon adenocarcinomas and metastatic deposits.32 In addition, it was possible to determine blindly the sex of the patients based on the methylation status of CpG islands on the inactivated copy of the X chromosome, a pattern observed in women only.33 Overall, this analysis provides evidence of the quality of DNA extracted and its compatibility with the methylation platform.
MicroRNA profiling
Total RNA extracted from patient biopsy samples were subjected to TaqMan-based quantitative microRNA profiling. As shown in Figure 4c, it was possible to identify reliably 138 microRNAs in all three samples. We then investigated whether the microRNA profiles obtained were consistent with previous reports in colorectal cancer profiling studies. Of 25 microRNA reported to be upregulated in colorectal cancer, 12 were also identified in our samples, including miR-(21, 26b, 132, 135, 141, 142, 181, 200, 203, 320, 372) and let-7b.34, 35 Interestingly, miR-21, which was detected in all three patient samples, is the microRNA most frequently upregulated in colorectal cancer and is known to target PTEN.35, 36
Splicing isoforms
Alternative splicing events were analyzed by high-throughput PCR.16 We selected a panel of 96 alternative splicing events based on their active splicing profiles in 11 cell lines derived from breast, ovary, uterus, prostate and colon (HCT-116) (unpublished data, http://palace.lgfus.ca/data/related/1166). By definition, an active alternative splicing event yields evidence of two or more isoforms in the sample set under consideration. Active ASEs are highly sensitive to changes in cellular conditions, and as such show strong promise as cancer-specific biomarkers.16 Figure 4d shows a heatmap representation of the splicing profiles of the active ASE test set. Changes in the regulation of specific ASEs can be detected between samples, represented by color changes in the heatmap. Analysis of data, representing the ratio of two isoforms derived from each ASE, indicated that 78 of the 96 ASEs are active in all three samples of this set (Figure 4d), confirming the suitability of the sample extraction and amplification protocol.
Gene expression
Global gene expression profiling was performed using Agilent Human whole-genome 60K arrays. The Agilent spike-in concentration–response statistics were in the manufacturer ranges with R2 values over 0.99 and slopes of 1.08, 1.10 and 1.07 for patients 003, 009 and 027, respectively, indicating that the technical processing of the arrays was carried out correctly without artifact. Figure 4e displays the 12 999 genes detected in all three patient samples. There is a good correlation between the expression level of different genes in the heatmap for all three patients with Pearson’s correlation coefficients ranging from 0.88 to 0.91 for all pairwise correlations between the three samples (all P-values <2.2e−16).
Integrated Analysis of the Genomic Data
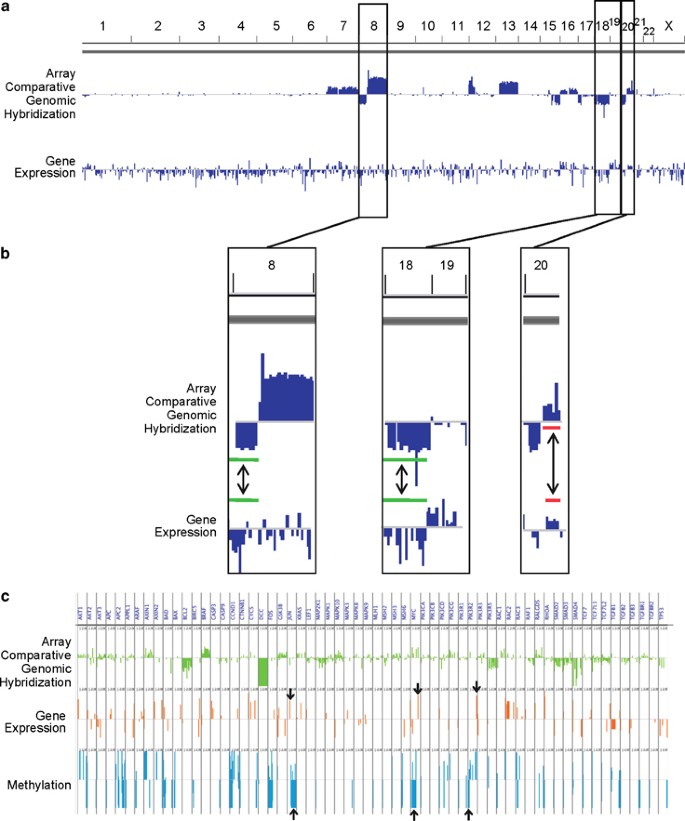
A comprehensive evaluation of a tumor’s molecular profile requires the integration of genomic data at multiple levels.37, 38 As a proof of concept, we used an integrative approach to show concordance between DNA and RNA genomic data obtained from patient liver biopsies. Figure 5 displays the result of integrating the array comparative genomic hybridization, gene expression and methylation data for patient 003. As expected, for deleted chromosomal regions (chromosomes 8 and 18) detected by array comparative genomic hybridization, there was a global downregulation of genes within the same region with significant difference with the distribution of gene expression from genes outside deleted regions (Figure 5b) (Wilcoxon’s signed-rank test P=2.95e−06 and P=4.58e−11 for chromosomes 8 and 18, respectively). For amplified regions, we can observe in region 20q (Figure 5b) a good concordance between amplification and increased gene expression (Wilcoxon’s signed-rank test P=1.1e−08). As shown in Figure 5c, we were able to ascertain upregulation of genes typically found to be activated in colon adenocarcinoma, including JUN, PIK3R2 and MYC that are, respectively, in the top 27, 20 and 7% most abundant genes detected on the array.39, 40 The gene expression data correlated well with hypomethylation of the proto-oncogenes (JUN, MYC) or promoter region in the case of PIK3R2. The concordance of results from the different platforms supports the compatibility of our next-generation biobanking protocol.
Discussion
The integrity of the initial material used for profiling is a major determinant of the validity of bioanalytical data.7, 10, 41 We have optimized a preanalytical workflow to isolate high-quality RNA and DNA simultaneously from a needle core biopsy of a metastatic liver lesion, which meets the technical demands of array comparative genomic hybridization, methylation profiling, microRNA profiling, splicing variant analysis and gene expression profiling. The use of RNAlater in our method has several advantages; it is non-toxic, inexpensive, stabilizes RNA and sustains RNA integrity for several days. A washing step before OCT embedding enables sectioning and tissue preservation for histological assessment, as has previously been demonstrated for breast specimens.20 While we demonstrate that OCT does not interfere with RNA quality and downstream enzymatic reactions, we recommend removing excess OCT since it saturates the Qiagen column and can decrease yield.
Embedding the biopsy in support media is needed to enable sectioning for the analysis of tumor content, the definition of which must be agreed upon by the biobanking pathologists. We demonstrate that tumor content varies significantly, even between successive biopsies obtained from the same lesion. In reports of multiplex analyses from patient tissue, this important preanalytical variable must be controlled as contributions from non-neoplastic cells can skew experimental results.8, 9 Measuring the percentage of neoplastic cells, necrosis and stroma in the tumor-containing region of the biopsy is semiquantitative at best and subject to interpretation, so centralizing histological analysis reduces variability. In the Q-CROC-01 study, two patients were subsequently diagnosed with neuroendocrine tumors, despite a primary diagnosis of colorectal cancer. Hence, histological analysis also serves to identify patients whose biopsies were not histologically compatible with their primary diagnosis.
An important consideration was that we obtain sufficient DNA and RNA from the biopsies to enable downstream multiplex analyses. Thus, laser microdissection was not a practical alternative. To safeguard against insufficient material, the collection of additional biopsies would be preferred. However, there is a delicate balance between increasing the risk to patients by performing multiple passes, and obtaining maximum yield of genomic material. We demonstrate that simultaneous extraction of DNA and RNA from a single core yields nucleic acids of optimal concentration and quality for our selected downstream genomic applications. We could detect a significant number of miRNAs in our biopsies without modifying the protocol to further enrich for miRNAs. However, with the use of an improved kit (AllPrep Universal) recently released by Qiagen, we notice further enrichment for miRNAs after Bioanalyzer analysis compared with the Allprep kit (data not shown). The tissue of origin is a determining factor for the development of standard operating procedures. As the liver is a common organ of metastatic deposits, this protocol could be adopted for sample manipulation and processing of liver metastatic lesions arising from cancers other than colorectal. Our experience with tissue processing for two other studies—lymph nodes for Q-CROC-02 (NCT01238692) and breast for Q-CROC-03 (NCT01276899) is that tissue-specific methodology must be developed (unpublished results). For example, tissue architecture and morphology is perfectly preserved when breast needle core biopsies are collected in RNAlater and frozen (protocol D, unpublished results), but here we show that this procedure cannot be applied to liver biopsies.
Next-generation biobanking is a translational research endeavour that requires a detailed workflow across various disciplines.42, 43 The expertise of interventional radiologists, pathologists and research technicians greatly affects the quality and usability of patient specimens. Ideally, a central lab and designated technician would systematically process these fragile samples. Alternatives to minimize preanalytical variability are that highly trained personnel follow strict protocols and/or that telepathology be used for central reviews. We agree with the BRISQ guidelines that a collective effort be made to report preanalytical variables. Furthermore, sample quality should be uniformly assessed so that translational discovery efforts can benefit from combining multiplex data across next-generation biobanks.
Although specific analytic technologies continue to advance rapidly, next-generation biobanking protocols must also evolve to minimize variability inherent to multicenter trials and improve data integration. However, protocols for sample procurement and processing are difficult to standardize as these are largely dependent on the tissue type, the demands of the analytical platforms, feasibility at different centers and cost. The protocol described here is being reviewed by the Worldwide Innovative Network Consortium for its first international biopsy-driven clinical trial, a study to select rational therapeutics based on the analysis of matched tumor and normal biopsies in patients with advanced malignancies. This will provide an important opportunity to further validate the utility of our approach in a wider range of tissues.
References
- Sequist LV, Waltman BA, Dias-Santagata D et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26.Article PubMed PubMed Central Google Scholar
- Nazarian R, Shi H, Wang Q et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010;468:973–977.CAS Article PubMed PubMed Central Google Scholar
- Vignot S, Besse B, André F et al. Discrepancies between primary tumor and metastasis: a literature review on clinically established biomarkers. Crit Rev Oncol Hematol 2012;84:301–313.Article PubMed PubMed Central Google Scholar
- Kim E, Herbst R, Wistuba I et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Disc 2011;1:44–53.CAS Article Google Scholar
- Barker AD, Sigman CC, Kelloff GJ et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther 2009;86:97–100.CAS Article PubMed Google Scholar
- Moore HM, Kelly A, Jewell SD et al. Biospecimen Reporting for Improved Study Quality. Biopreserv Biobank 2011;9:57–70.Article PubMed PubMed Central Google Scholar
- Simeon-Dubach D, Burt AD, Hall PA . Quality really matters: the need to improve specimen quality in biomedical research. J Pathol 2012;228:431–433.Article PubMed Google Scholar
- Cleator SJ, Powles TJ, Dexter T et al. The effect of the stromal component of breast tumours on prediction of clinical outcome using gene expression microarray analysis. Breast Cancer Res 2006;8:R32.Article PubMed PubMed Central Google Scholar
- Nelson PT, Wang WX, Wilfred BR et al. Technical variables in high-throughput miRNA expression profiling: much work remains to be done. Biochim Biophys Acta 2008;1779:758–765.CAS Article PubMed PubMed Central Google Scholar
- Verderio P . Assessing the clinical relevance of oncogenic pathways in neoadjuvant breast cancer. J Clin Oncol 2012;30:1912–1915.Article PubMed Google Scholar
- Begley CG, Ellis LM . Drug development: raise standards for preclinical cancer research. Nature 2012;483:531–533.CAS Article Google Scholar
- Patel IJ, Davidson JC, Nikolic B et al. Consensus guidelines for periprocedural management of coagulation status and hemostasis risk in percutaneous image-guided interventions. J Vasc Interv Radiol 2012;23:727–736.Article PubMed Google Scholar
- Przybytkowski E, Ferrario C, Basik M . The use of ultra-dense array CGH analysis for the discovery of micro-copy number alterations and gene fusions in the cancer genome. BMC Med Genom 2011;4:16.CAS Article Google Scholar
- Smyth GK . Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3.Article PubMed Google Scholar
- Thorvaldsdóttir H, Robinson JT, Mesirov JP . Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2012;14:178–192.Article PubMed PubMed Central Google Scholar
- Venables JP, Klinck R, Koh C et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol 2009;16:670–676.CAS Article PubMed Google Scholar
- Venables JP, Klinck R, Bramard A et al. Identification of alternative splicing markers for breast cancer. Cancer Res 2008;68:9525–9531.CAS Article PubMed Google Scholar
- Vandesompele J, De Preter K, Pattyn F et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034.Article PubMed PubMed Central Google Scholar
- Hoffmann G, Ijzer J, Brinkhof B et al. Comparison of different methods to obtain and store liver biopsies for molecular and histological research. Comp Hepatol 2009;8:3.Article PubMed PubMed Central Google Scholar
- Ellis M, Davis N, Coop A et al. Development and validation of a method for using breast core needle biopsies for gene expression microarray analyses. Clin Cancer Res 2002;8:1155–1166.CAS PubMed Google Scholar
- Srinivasan M, Sedmak D, Jewell S . Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 2002;161:1961–1971.CAS Article PubMed PubMed Central Google Scholar
- Network TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337.Article Google Scholar
- Camps J, Armengol G, del Rey J et al. Genome-wide differences between microsatellite stable and unstable colorectal tumors. Carcinogenesis 2006;27:419–428.CAS Article PubMed Google Scholar
- Nakao K, Mehta KR, Fridlyand J et al. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis 2004;25:1345–1357.CAS Article PubMed Google Scholar
- Douglas EJ, Fiegler H, Rowan A et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res 2004;64:4817–4825.CAS Article PubMed Google Scholar
- Kurashina K, Yamashita Y, Ueno T et al. Chromosome copy number analysis in screening for prognosis-related genomic regions in colorectal carcinoma. Cancer Sci 2008;99:1835–1840.CAS Article PubMed Google Scholar
- Hermsen M, Postma C, Baak J et al. Colorectal adenoma to carcinoma progression follows multiple pathways of chromosomal instability. Gastroenterology 2002;123:1109–1119.CAS Article PubMed Google Scholar
- De Angelis PM, Clausen OP, Schjølberg A et al. Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br J Cancer 1999;80:526–535.CAS Article PubMed Google Scholar
- Gama-Sosa MA, Slagel VA, Trewyn RW et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 1983;11:6883–6894.CAS Article PubMed PubMed Central Google Scholar
- Goelz SE, Vogelstein B, Hamilton SR et al. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 1985;228:187–190.CAS Article Google Scholar
- Kanehisa M, Goto S, Furumichi M et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res 2010;38:D355–D360.CAS Article PubMed Google Scholar
- Sharrard RM, Royds JA, Rogers S et al. Patterns of methylation of the c-myc gene in human colorectal cancer progression. Br J Cancer 1992;65:667–672.CAS Article PubMed PubMed Central Google Scholar
- Hellman A, Chess A . Gene body-specific methylation on the active X chromosome. Science 2007;315:1141–1143.CAS Article PubMed Google Scholar
- Yang L, Belaguli N, Berger DH . MicroRNA and colorectal cancer. World J Surg 2009;33:638–646.Article PubMed Google Scholar
- Schetter AJ, Leung SY, Sohn JJ et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008;299:425–436.CAS PubMed PubMed Central Google Scholar
- Slaby O, Svoboda M, Michalek J et al. MicroRNAs in colorectal cancer: translation of molecular biology into clinical application. Mol Cancer 2009;8:102.Article PubMed PubMed Central Google Scholar
- Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–615.Article Google Scholar
- Taylor BS, Schultz N, Hieronymus H et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010;18:11–22.CAS Article PubMed PubMed Central Google Scholar
- Magrisso IJ, Richmond RE, Carter JH et al. Immunohistochemical detection of RAS, JUN, FOS, and p53 oncoprotein expression in human colorectal adenomas and carcinomas. Lab Invest 1993;69:674–681.CAS PubMed Google Scholar
- Guo C, Sah JF, Beard L et al. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer 2008;47:939–946.CAS Article PubMed PubMed Central Google Scholar
- Kissinger CB, Kissinger PT . Your bioanalytical data are only as good as your samples. Bioanalysis 2012;4:1411–1415.CAS Article PubMed Google Scholar
- Aguilar-Mahecha A, Diaz Z, Buchanan M et al. Making personalized medicine a reality: the challenges of a modern translational research biopsy-driven program in an academic setting: the Segal Cancer Center experience. J Med Person 2011;9:8.Article Google Scholar
- Meric-Bernstam F, Mills GB . Overcoming implementation challenges of personalized cancer therapy. Nat Rev Clin Oncol 2012;9:542–548.CAS Article PubMed Google Scholar
Acknowledgements
We acknowledge the patients, site coordinators and interventional radiologists at the centers participating in the Q-CROC-01 study. We are grateful to Caroline Pouliot for producing artwork. The Q-CROC-01 trial is supported by a PFIZER-FRSQ Innovation Fund award and by Sanofi-Aventis. ERP is supported by a Doctoral scholarship from Canadian Institutes of Health Research (CIHR). The funding organizations have no role in study design, data collection and analysis, approval or preparation of the manuscript.
Author information
Author notes
- Zuanel Diaz, Adriana Aguilar-Mahecha and Eric R Paquet: These authors contributed equally to this work.
Affiliations
- Quebec Clinical Research Organization in Cancer, Montreal, QC, CanadaZuanel Diaz, Thérèse Gagnon-Kugler, Samia Qureshi, Caroline Rousseau & Gerald Batist
- Department of Oncology, Jewish General Hospital, Segal Cancer Center, McGill University, Montreal, QC, CanadaAdriana Aguilar-Mahecha, Mark Basik, Petr Kavan, Cathy Lan, Catherine Chabot, Marguerite Buchanan, Ewa Przybytkowski & Gerald Batist
- Department of Biochemistry, McGill University, Montreal, QC, CanadaEric R Paquet
- Department of Pathology, Hôpital du Saint-Sacrement, Quebec, QC, CanadaMichèle Orain & Bernard Têtu
- Department of Radiology, Jewish General Hospital, Montreal, QC, CanadaErrol Camlioglu & André Constantin
- Department of Pathology, Jewish General Hospital, Montreal, QC, CanadaNaciba Benlimame, Adrian Gologan & Alan Spatz
- Department of Molecular Medicine, Laval University, Quebec, QC, CanadaDimcho Bachvarov
- Laval University Cancer Research Center, Hôtel-Dieu de Québec (CHUQ), Quebec, QC, CanadaGuillaume Jannot & Martin J Simard
- Département de microbiologie et d’infectiologie, Université de Sherbrooke, Faculté de médecine et des sciences de la santé, Sherbrooke, QC, CanadaBenoit Chabot & Roscoe Klinck
- Department of Medicine, Hôpital du Sacré-Cœur de Montréal, Montreal, QC, CanadaBernard Lespérance
- Department of Oncology, Hôpital Charles Lemoyne, CICM, Greenfield Park, QC, CanadaBenoit Samson
- Department of Oncology, McGill University Health Center, Montreal, QC, CanadaPetr Kavan & Thierry Alcindor
- Department of Medical Oncology, St Mary’s Hospital Center, Montreal, QC, CanadaRichard Dalfen
Corresponding author
Correspondence to Gerald Batist.
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Supplementary Information (DOC 31 kb)
Rights and permissions
About this article
Cite this article
Diaz, Z., Aguilar-Mahecha, A., Paquet, E. et al. Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine. Mod Pathol 26, 1413–1424 (2013). https://doi.org/10.1038/modpathol.2013.81
- Received18 January 2013
- Revised25 February 2013
- Accepted27 February 2013
- Published07 June 2013
- Issue DateNovember 2013
- DOIhttps://doi.org/10.1038/modpathol.2013.81